INDIVIDUAL-LEVEL DATA ACCESS INFORMATION
Last updated: 16 June 2025
*Note: this webpage covers individual-level/ restricted access data. PGC GWAS summary statistics are available without restriction on the Download Results webpage.
The PGC has established a large collection of individual-level genetic and phenotypic data, which is accessible to investigators for Secondary Analysis Proposals, via application to the relevant PGC Workgroup(s). These data are stored on the Snellius server hosted by SURFsara which is located in the Netherlands. The PGC is committed to data sharing to engage more research groups, maximize knowledge, and accelerate scientific progress, while respecting the limits of national laws and ethical restrictions. Here, we provide an overview of the individual-level data available, the application process, and answers to some FAQs. The data access infrastructure is managed by the PGC Data Access Committee, which includes representatives from each PGC Disorder Workgroup.
Data available
- At present, 11 PGC Disorder Workgroups have collections of individual-level genetic data per cohort stored on the Snellius server and available for Secondary Analysis Proposals. Permission to access to each disorder collection is granted by the relevant PGC Disorder Workgroup.
- Genetic data available per cohort includes raw genotype data, QC’ed genotype data, imputed dosage data, imputed dosages converted to hard calls, and principal components of genetic ancestry. Files are provided in RICOPILI format. Some Disorder Workgroups have additional types of genetic data available, such as pre-calculated polygenic risk scores, imputed HLA or C4 data.
- Phenotypic data available for all cohorts includes case-control status. Some Disorder Workgroups have more detailed subphenotype information available.
- Within each PGC disorder, the data for most cohorts are accessible through a “fast-track” data package, which is accessible once the Workgroup approves the Secondary Analysis Proposal. However, some cohorts require explicit permission from the Principal Investigator of the cohort or from a data repository such as dbGAP. If investigators wish to access these cohorts on Snellius, they will need to secure the special permissions required.
- The latest GWAS publication from the PGC Disorder Workgroup typically contains information about the specific cohorts and sample sizes available in the Supplementary Materials. For specific questions about the individual-level data available on Snellius and the permissions required, please contact the relevant PGC Disorder Workgroup Chair(s) and the Data Access Committee Representative (DAC Rep).
Overview of data application process
Here PGC Data Access Committee Co-chair Lea Davis describes the process in a YouTube video.
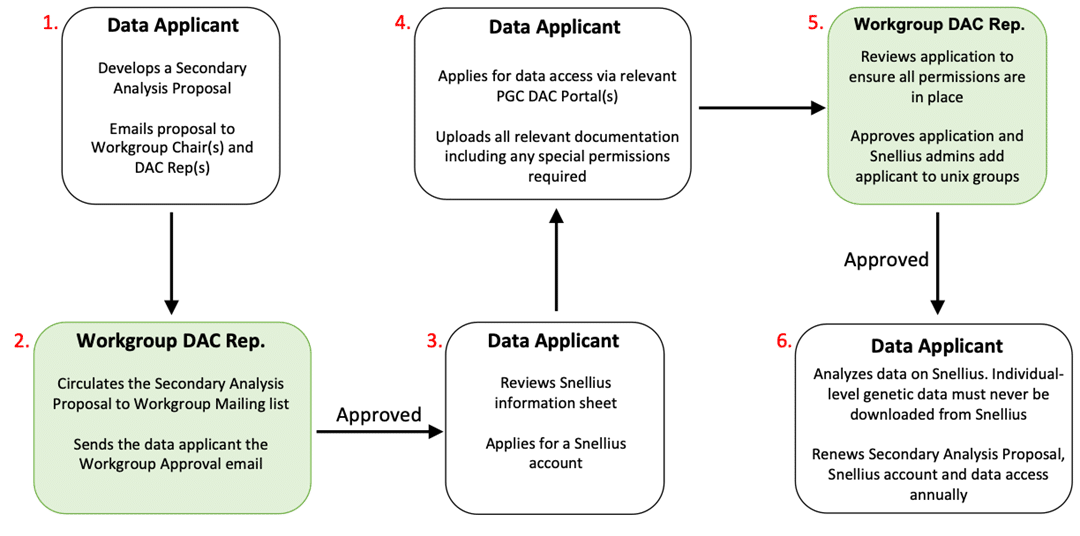
- The applicant develops a Secondary Analysis Proposal and submits it to the relevant PGC Workgroup(s), by emailing it to the Workgroup Chair(s) and Data Access Committee (DAC) Representative(s). This includes the study rationale, analytic plans, data being requested, individuals involved in the work, timeline, and publication plans. Please ensure sufficient information is included for members of the Workgroup to review the proposed project.
- The Secondary Analysis Proposal will be circulated to the Workgroup mailing list for review by all members. The Workgroup Chair(s) and DAC Representative(s) will advise on whether a short presentation of the Secondary Analysis Proposal is required on a Workgroup call. Once the Secondary Analysis Proposal is approved by the Workgroup(s), the Workgroup Chair(s) or DAC Representative(s) will send you an approval email.
- After the Secondary Analysis Proposal is approved, the analyst(s) named on the proposal will obtain an account on the Snellius server, where the individual-level data are stored. Prior to applying for a Snellius account, please review this informational sheet. To obtain a Snellius account, follow these instructions. Approval typically takes a few days.
- Apply for data access via the relevant PGC Data Access Portal(s). You will need your Snellius username, and several documents such as a copy of your Secondary Analysis Proposal, a copy of your Workgroup approval email and documentation for any specific permissions required such as dbGaP. Detailed information about all of the documents required is provided below.
- Your data access request through the portal will be reviewed by the relevant DAC Representative(s) and once approved the Snellius administrators will add your username to the relevant unix permissions groups. Note: Individual-level genetic data may never be downloaded from Snellius.
- Secondary Analysis Proposals, Snellius accounts and data access are approved for one year at a time and you must renew all three of these annually. You will receive reminder emails one month before these are due to expire.
Acknowledgements
The PGC is deeply grateful to SURFsara and VU University Amsterdam for their support in using the Dutch National Supercomputer Snellius.
Documents required for data access
Always required:
- Secondary Analysis Proposal - Please use the template provided and ensure sufficient information is included for members of the Workgroup to review the proposed project and make a recommendation.
- Workgroup Approval Email(s) - This will be provided to you by the Workgroup Chair(s) or Data Access Committee (DAC) Representative(s)
- Signed PGC Analyst Memo - We request that everyone who accesses individual-level PGC data (and their supervisor if applicable) confirms their agreement with the PGC Memorandum of Understanding (MOU) and understands the obligations and benefits of participating. Please read the MOU carefully and follow all of the steps.
- Signed WTCCC Data Access Agreement - This is required for accessing cohorts from the Wellcome Trust Case Control Consortium.
May be required depending on the cohorts you are applying for:
- Current dbGaP Approval - Some PGC cohorts were submitted to the NIH dbGaP Repository by their Principal Investigators and have been gathered together into the PGC dbGaP collection. The PGC can only provide access to these cohorts on Snellius to investigators with an approved and currently valid dbGaP application for these data. This means there’s no need for you to download the data from dbGaP and format the files yourself. This dbGaP application can be used as a template for your own. Note, the Principal Investigator of the lab should apply, as their application covers everyone who reports to them. Multiple PIs at the same institute can be added as collaborators on one dbGaP application. Collaborators at different institutes need their own dbGaP application, since each application must be signed by an Institutional Signing Official. Your approval from dbGaP is valid for one year and it must be renewed via dbGaP annually. You must upload a valid dbGaP approval via the PGC DAC portal annually to maintain access to the cohorts in the PGC dbGaP collection on Snellius.
- PI approval emails - Some PGC cohorts require explicit permission from the Principal Investigator of the cohort for access. You will need a copy of the approval email from the cohort PI to access these.
- HRC reference panel approval - Most PGC datasets are imputed to the Haplotype Reference Consortium (HRC) reference panel, you need HRC access to run GWAS using RICOPILI, and we use it as a reference panel for many post-GWAS analyses. The PGC can provide access to the RICOPILI-formatted version of the HRC on Snellius to investigators with approved HRC access applications from the Sanger institute. Here you can request access to the HRC using its EGA Dataset (EGAD) identifier EGAD00001002729. Each individual who accesses the data needs to be named on this application, so you may want to add names of your trainees at your institute. It's possible to add people on later by emailing datasharing@sanger.ac.uk.
FAQ
Are there authorship requirements for using individual-level PGC data?
Yes, requirements may include individual named authorship for a number of investigators from each cohort analyzed or including a PGC Disorder Workgroup consortium byline, depending on your project. PGC Disorder Workgroups typically have an authorship policy for different types of manuscripts which is available on request from the Chair(s) of the relevant PGC Workgroup(s).
How long will it take to have my Secondary Analysis Proposal reviewed by the relevant PGC Workgroup(s)?
Typically Secondary Analysis Proposals are reviewed within three weeks. It may take longer if the Workgroup requires a short presentation of the Secondary Analysis Proposal on a Workgroup call, as these typically occur monthly.
Once my Secondary Analysis Proposal is approved, can I expand my project or use the data for another project?
No, Secondary Analysis Proposals are only for the specific project described. If you wish to use the data for another project, you will need to submit another Secondary Analysis Proposal to the relevant PGC Disorder Workgroup(s). We realize that analytic strategies can sometimes change through the course of analysis. If you find that your plan has significantly shifted course, you should speak with your Data Access Committee Representative (DAC Rep) to determine if the project has changed enough to be considered a new project.
I have specific questions about the process; who do I contact?
Please contact the Data Access Committee Representative (DAC Rep) from the relevant PGC Disorder Workgroup(s).
