Join the PGC
Last updated: 16 June 2025
The PGC accepts both individual-level genetic and phenotypic data and GWAS summary statistics. You can share your data with one or more PGC disorder workgroups, and data are stored on a secure computing server “Snellius” located in the Netherlands. We have developed a standardized process to streamline the submission of data and welcome new members to the PGC. Here, we provide an overview of the submission process, instructions on how to get started, and answers to some FAQs. This entire process is managed by the PGC Data Receiving Committee, which includes representatives from each PGC disorder workgroup.

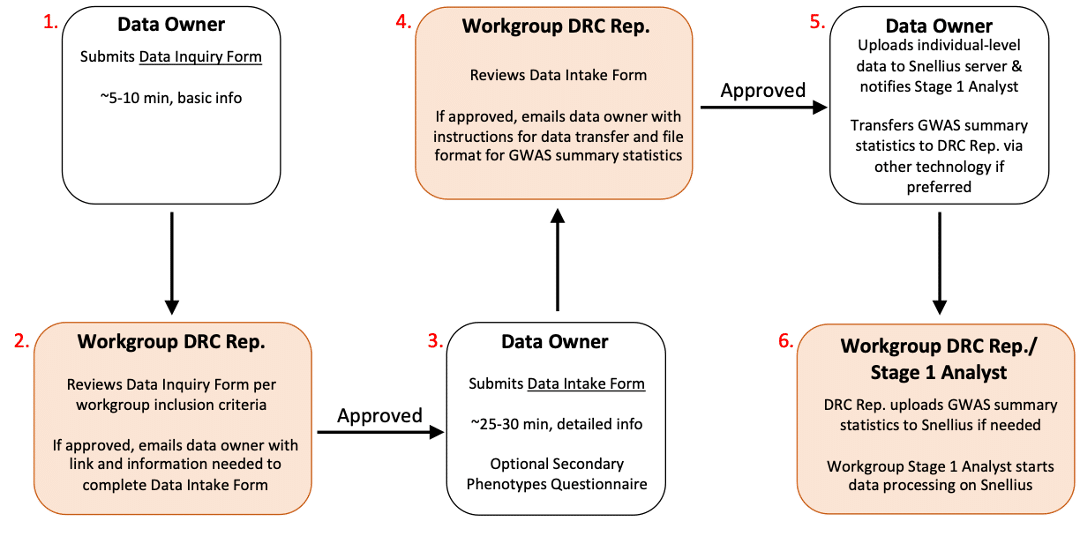
Overview of data submission process
Here PGC Data Receiving Committee Co-chair Lea Davis describes the process in a YouTube video.
- The data owner submits the online PGC Data Inquiry Form (takes 5-10 mins) which collects basic information about the cohort, and the PGC Workgroup(s) you’re interested in sharing data with.
- The Data Receiving Committee Representative(s) (DRC Rep) from the relevant PGC Workgroup(s) will review your PGC Data Inquiry Form, to confirm the cohort meets their inclusion criteria. Once the DRC Rep approves your PGC Data Inquiry Form, you’ll receive an automated email including a link to the online PGC Data Intake Form and instructions on the information you’ll need to prepare to complete this form.
- The data owner completes the PGC Data Intake Form (takes 20-25 mins) which collects detailed information about the cohort, including a publication-ready cohort description, ethics information, and analysis details in the case of GWAS summary statistics. This form also includes an optional Secondary Phenotypes Questionnaire.
- The DRC Rep(s) will review your PGC Data Intake Form and once it’s approved, you’ll receive an email with instructions for data transfer and file formatting requirements in the case of GWAS summary statistics.
- The data owner uploads individual-level data to the Snellius server for data security, and emails the Workgroup the Stage 1 analyst to notify them of the data upload and location once complete. GWAS summary statistics can be transferred to the DRC Rep(s) using other technologies if the data owner prefers.
- The DRC Rep(s) will upload GWAS summary statistics to Snellius if needed. The Stage 1 Analyst from the relevant PGC Disorder Workgroup(s) will start data processing on the Snellius server.
Get Started
This form takes 5-10 mins to complete. You will be asked to provide basic information that will help the Data Receiving Committee Representative (DRC Rep) determine whether your data meet the Workgroup’s inclusion criteria. You’ll receive a confirmation email with a copy of your submission and the DRC Rep(s) will be in touch soon.
FAQ
What types of individual-level genetic data are the PGC interested in receiving?
In general, the PGC is interested in receiving array-based genotyping data (e.g., vcf/ plink/ bgen files), intensity data for CNV calling (e.g., cel/ idat files), and phenotype data, including various secondary phenotypes. Some PGC Workgroups are also interested in sequence data or various types of ‘omics data. If you have a question about submitting certain types of data, please get in touch with the Data Receiving Committee Representative (DRC Rep) from the relevant PGC Disorder Workgroup.
What are the advantages of sharing individual-level data with the PGC?
The PGC will process your data from start to finish, including quality control, ancestry assignment, relatedness checks, imputation, CNV calling and analysis (if array intensity data are provided), using our established pipelines. We can provide a copy of the processed data back to you for storage on your local institutional server. When submitting data, you can specify whether you’d like to make your individual-level data available on the Snellius server for Secondary Analysis Proposals submitted by other investigators and any specific data access restrictions you may have. Secondary Analysis Proposals require a formal application to the relevant PGC Disorder Workgroup. More information on this process can be found on the PGC Data Access webpage.
Is there a minimum sample size requirement/ can I share controls only?
Each PGC Disorder Workgroup makes its own determination on the minimum number of samples required. Depending on the characteristics of your data, it may be possible to combine them with other datasets. The Data Receiving Committee Representative (DRC Rep) will assess each cohort based on the PGC Data Inquiry Form. The PGC may accept controls for potential matching with case-only datasets. As a rule of thumb, 50 individuals in a dataset serves as a minimum cohort size, though exceptions are possible.
Where will my data be stored?
Your data will be stored on the Snellius server hosted by SURFsara, located in the Netherlands. Snellius is GDPR-compliant, HIPAA-compliant and meets international cybersecurity standards (ISO 27001 certified, which is equivalent to U.S. NIST SP 800-171 certification). Individual-level genetic data shared with the PGC will not be downloaded from Snellius, stored anywhere else, or uploaded to websites such as imputation servers. Exceptions can be made in rare circumstances, with written permission from the data owner.
How do I transfer my data?
Individual-level data must be transferred by secure copy protocol (scp) to the Snellius server to ensure the security of your data. Once your PGC Data Intake Form is approved, you’ll receive an email with instructions on how to obtain a Snellius login and complete the data transfer. GWAS summary statistics can be shared with the DRC Rep using other technologies if you prefer and they will complete the upload to Snellius. You can always talk to the Stage 1 Analyst for alternative sharing possibilities.
What information will the PGC need about my data/ cohort?
The PGC Data Intake Form gathers a detailed referenced description of the cohort, information on case and control ascertainment, a file manifest (if available), and your IRB approved study protocol (if available). Once your PGC Data Inquiry Form is approved, you’ll receive an email with the link to the PGC Data Intake Form and the details about the information you’ll need to prepare to complete it.
Who will have access to my data?
The DRC Rep, Stage 1 Analyst and Stage 2 Analyst from the PGC Disorder Workgroup(s) you submit to will have access to your data. Some other analysts in these PGC Disorder Workgroup(s) may also access the data for post-GWAS analyses such as polygenic risk scoring (this is typically discussed on the disorder workgroup calls). When submitting data, you can specify whether you’d like to make your individual-level data available on the Snellius server for Secondary Analysis Proposals submitted by other investigators and any specific data access restrictions you may have. Secondary Analysis Proposals require a formal application to the relevant PGC Disorder Workgroup. More information on this process can be found on the PGC Data Access webpage.
How will my data be used by the PGC?
Your data will be included in primary analyses conducted by the PGC Disorder Workgroup(s) you submit your data to. When submitting data, you can specify whether you’d like to make your individual-level data available on the Snellius server for Secondary Analysis Proposals submitted by other investigators and any specific data access restrictions you may have. Secondary Analysis Proposals require a formal application to the relevant PGC Disorder Workgroup. More information on this process can be found on the PGC Data Access webpage.
How will I stay up-to-date on how my data are being used?
Investigators who share data are welcome to attend meetings and join the mailing list of the relevant PGC Disorder Workgroup(s). Please contact the Chair(s) of the relevant PGC Workgroup(s) to be added.
Will I get co-authorship for sharing my data?
Yes, investigators who share data with the PGC have the opportunity for co-authorship on publications which use their data. This may include individual named authorship, or being listed under the PGC Disorder Workgroup consortium byline, depending on how your data are used. PGC Disorder Workgroups typically have an authorship policy for different types of manuscripts which is available on request from the Chair(s) of the relevant PGC Workgroup(s). The number of co-authors can be negotiated with the Workgroup Chair(s) and typically reflects the sample size of your cohort.
Can I withdraw my data?
In the event that you need to withdraw data shared with the PGC, please contact the Chair(s) of the relevant PGC Workgroup(s) to notify them and discuss the next steps. Even though it is possible to remove genotypes from the server, published results will not be retracted naturally.
I have specific questions about the process; who do I contact?
Please contact the Data Receiving Committee Representative (DRC Rep) from the relevant PGC Disorder Workgroup(s).
